1. Introduction
rotein and other biomedical entity name are used in various biomedical and other bioinformaticsrelated research. So we will have to work hard to identifying the entities. In text-based literature protein and other biomedical name are tagged with other text. Identifying such entities from text file is very difficult. So we will have to use any scientific approach to solve the problem. Natural language processing is a system which can be used to solve the problem. Using natural language processing we will extract the required info from a text file. We use 'GENIA tagger' database to extract the information from the pdf file and get our required biomedical name. Then we will use these names to make a relation between them and visualized them.
2. II.
3. Related Work
Many researches are introduced in the field of biomedical and bioinformatics by using data extraction technique. All the work is currently done by text reading system. It is not possible to get the accurate data from manually text extraction technique. As a result protein and other Biomedical entities are not possible to find out correctly. So it is huge drawback of these types of a research field. In the research we have tried to find out the protein, and gene name which is about 70%-80% correct.
Author ? ? ?: e-mails: arif25-627@diu.edu.bd, Muhammad25-631@diu.edu.bd, saifuddin25-630@diu.edu.bd III.
4. Our Proposed Work
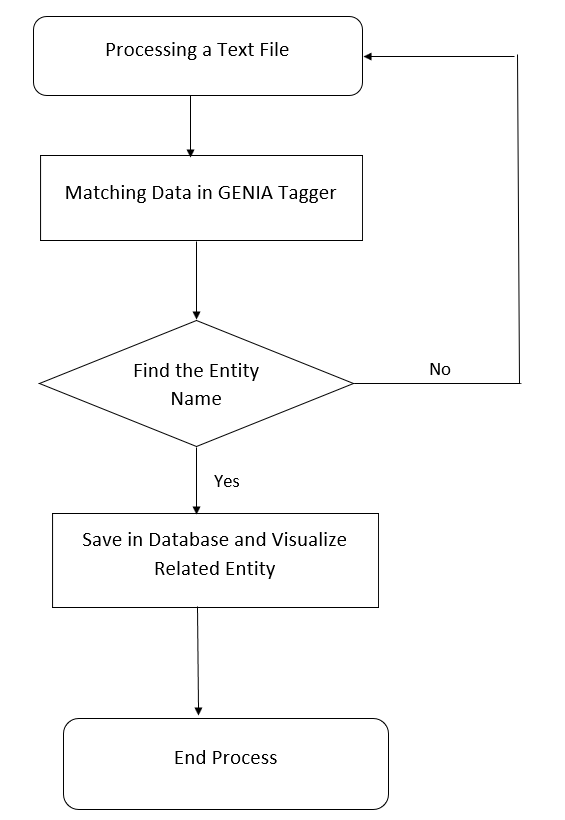
In the research we try to identify the tagging problem and find a solution related to this type of work. Our work will follow the below procedure. Pdf to text file conversion is looks complex task, but we can convert it easily with the use of the algorithm and other tools. For our project work, we will have to work on the natural language based text extraction system, which will identify which type of the data is in the text file. Moreover, we can find out the required data and another type of system approach to find that entity. The natural language based system will help us on the text file to find out the necessary information for the system fulfillment of the data. By using this system, we can find out protein and its related entity.
5. VI.
6. Using Genia Tagger for Data Extraction
GENIA tagger is a website that will help to find out the natural language based system for the protein name tagging from the text; we will use this website for the relevant data search. Moreover, we will use this information in the desired data analyze technique [5]. We also use these type of system for our data processing system [6].
7. VII.
8. Data Tagging
First we will keep the data in the text file. These data will help us in accessing the information [7] [8].
Protein name contains an acronym abbreviating the species name, e.g. Protein human growth hormone (hGH)/protein, but long-form human protein IGF-II / protein /long-form. Protein entities share common terms; there may be only one name entity that can be easily tagged. We tag such name as a protein. Long-form protein CSN subunits 4 /protein, 5,6 /long-form. Assessment of v2 the results on intercoder reliability using the revised guidelines are much better. Retrieving Data and Save Database According to the Related Entity
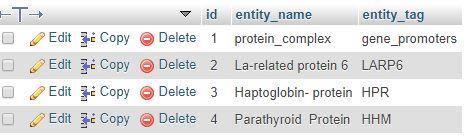
We will have to save data according to the text file which we will get from GENIA tagger website. Then we will able to visualize them.
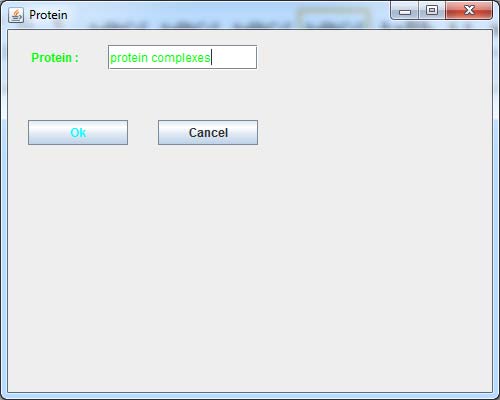
9. Data Visualization
After retrieving the data, we will analyze all the entities which are related to each other. We will categorize them according to the protein, gene, chromosome various entities. Then we will visualize protein name contains an acronym abbreviating the species name , e.g . Protein human growth hormone ( hGH ) /pro-tein , but long-form human protein IGF-II / protein /long-form . protein entities share common terms , there may be only one name entity that can be easily tagged . We tag such an entity as a protein, while the list of enti-ties together are tagged as a long-form, e.g . Long-form protein CSN subunits 4 /protein, 5, 6 /long-form . Assessment of v2 The results on inter-coder reliability using the revised guidelines are much better . We present results for F-measure
10. Named Entity Recognition Performance
Our pdf file contains lots of entity of Protein, DNA, RNA, Cell Line, and Cell Type. Genia tagger provides us the flowing the final performance on the evaluation set is as follows [12].
11. Conclusion
Natural Language Processing is a way to find out the similar relational data from a text or document. We try find out related protein and other biomedical entity name and visualize them. Our research makes the system fruitful for the data analysis process.
12. Entity